[PYTHON] Non-negative Matrix Factorization (NMF) with scikit-learn
NMF is a dimension reduction method and is said to be able to improve the accuracy of recommendations.
You can also easily use NMF with the Opportunity Learning Library scikit-learn.
This time, the purpose is to try it with concrete examples and understand NMF intuitively.
The article What is Matrix Factorization is easy to understand.
What is NMF
To get a rough idea of Matrix Factorization as a recommendation, it's in English, but the following materials are easy to understand. Matrix Factorization Techniques For Recommender Systems
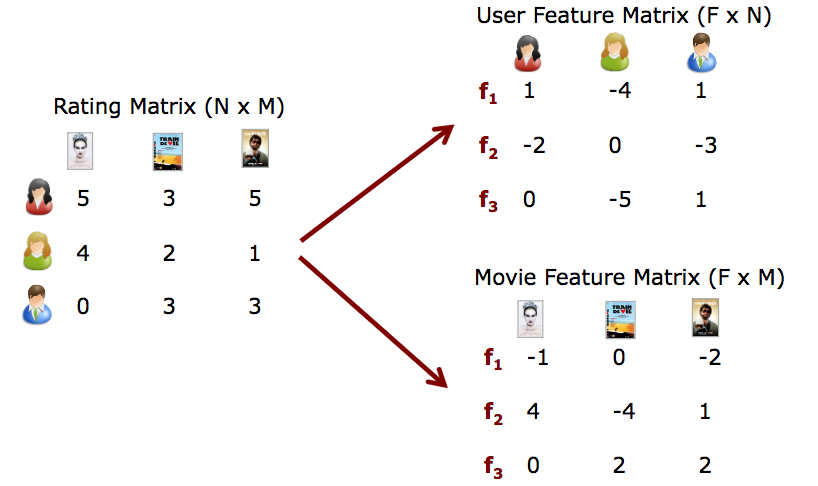
I will briefly explain the image by quoting the figure on this slide. Suppose you're assuming the Netflix Prize, where the NMF method has become so popular, and you're given a matrix that shows how many points a user and that person gave to which video (the Matrix on the left). If the user rated the video, it was rated from 1 to 5, and if it was not rated, 0 was entered. The fact that 0 is included is the heart of this problem, and we predict the true value of this 0. This Rating Matrix is decomposed into two matrices, "feature x user" and "feature x movie". Roughly speaking, NMF is a method for successful matrix factorization.
NMF formulation
\mathbf{R} \approx \mathbf{P} \times \mathbf{Q}^T = \hat{\mathbf{R}}
The error can be expressed as $ e_ {ij} ^ 2 $.
e_{ij}^2 = (r_{ij} - \hat{r}_{ij})^2 = (r_{ij} - \sum_{k=1}^K{p_{ik}q_{kj}})^2
However, the approximated $ \ hat {\ mathbf {R}} $ elements are as follows:
\hat{r}_{ij} = p_i^T q_j = \sum_{k=1}^k{p_{ik}q_{kj}}
0.5 * ||X - WH||_Fro^2
+ alpha * l1\_ratio * ||vec(W)||_1
+ alpha * l1\_ratio * ||vec(H)||_1
+ 0.5 * alpha * (1 - l1\_ratio) * ||W||_Fro^2
+ 0.5 * alpha * (1 - l1\_ratio) * ||H||_Fro^2
||A||_Fro^2 = \sum_{i,j} A_{ij}^2 (Frobenius norm)
||vec(A)||_1 = \sum_{i,j} abs(A_{ij}) (Elementwise L1 norm)
Implementation with scikit-learn
It's easy to call. For example, the code when the rating matrix is R and the feature dimension is 2 is as follows.
from sklearn.decomposition import NMF
model = NMF(n_components=2, init='random', random_state=0) # n_Specify the feature dimension with components
P = model.fit_transform(R) #Learning
Q = model.components_
Concrete example
Suppose R is a 5x4 matrix like this: The numbers in this matrix are used on this site. I referred to the example.
| D1 | D2 | D3 | D4 | |
|---|---|---|---|---|
| U1 | 5 | 3 | - | 1 |
| U2 | 4 | - | - | 1 |
| U3 | 1 | 1 | - | 5 |
| U4 | 1 | - | - | 4 |
| U5 | - | 1 | 5 | 4 |
from sklearn.decomposition import NMF
import numpy as np
#The 0 part is unknown
R = np.array([
[5, 3, 0, 1],
[4, 0, 0, 1],
[1, 1, 0, 5],
[1, 0, 0, 4],
[0, 1, 5, 4],
]
)
#Try changing the feature dimension k from 1 to 3
for k in range(1,4):
model = NMF(n_components=k, init='random', random_state=0)
P = model.fit_transform(R)
Q = model.components_
print("****************************")
print("k:",k)
print("P is")
print(P)
print("Q^T is")
print(Q)
print("P×Q^T is")
print(np.dot(P,Q))
print("R-P×Q^T is")
print(model.reconstruction_err_ )
The output result is as follows.
****************************
k: 1
P is
[[ 0.95446553]
[ 0.64922245]
[ 1.12694515]
[ 0.87376625]
[ 1.18572187]]
Q^T is
[[ 1.96356213 1.0846958 1.24239928 3.24322704]]
P×Q^T is
[[ 1.87415238 1.03530476 1.18582729 3.09554842]
[ 1.27478862 0.70420887 0.8065935 2.10557581]
[ 2.21282681 1.22239267 1.40011583 3.65493896]
[ 1.71569432 0.94777058 1.08556656 2.83382232]
[ 2.32823857 1.28614754 1.47314 3.84556523]]
R-P×Q^T is
7.511859871919941
****************************
k: 2
P is
[[ 0. 1.69547254]
[ 0. 1.13044637]
[ 1.38593123 0.42353044]
[ 1.08595617 0.3165454 ]
[ 2.01891156 0. ]]
Q^T is
[[ 0. 0.32199795 1.40668938 2.58501889]
[ 3.09992487 1.17556787 0. 0.85823043]]
P×Q^T is
[[ 5.25583751 1.99314304 0. 1.45510614]
[ 3.50429883 1.32891643 0. 0.97018348]
[ 1.31291255 0.9441558 1.94957474 3.94614513]
[ 0.98126695 0.72179626 1.52760301 3.0788861 ]
[ 0. 0.65008539 2.83998144 5.21892451]]
R-P×Q^T is
4.2765298341191516
****************************
k: 3
P is
[[ 0.08750151 2.03662182 0.52066139]
[ 0. 1.32090927 0.59992585]
[ 0.04491053 0.44753619 3.4215759 ]
[ 0. 0.3221337 2.75625987]
[ 2.6249395 0. 0.91559966]]
Q^T is
[[ 0. 0.38636917 1.90213535 1.02176621]
[ 2.62017626 1.02934221 0. 0.08629708]
[ 0. 0.02388804 0. 1.43875694]]
P×Q^T is
[[ 5.33630814 2.14262626 0.16643971 1.0142658 ]
[ 3.4610151 1.37399871 0. 0.97713811]
[ 1.17262369 0.55975468 0.08542591 5.00732522]
[ 0.84404708 0.39742747 0. 3.99338722]
[ 0. 1.03606758 4.99299022 3.99939987]]
R-P×Q^T is
1.8626938982246695
The observation matrix R was a 5 × 4 matrix. When you try different k, you can see that P is a 5 × k matrix and Q ^ T is a k × 4 matrix.
Also, in P × Q ^ T, it is estimated that it was 0 in R. For example, when k = 2, the following $ \ hat {\ mathbf {R}} $ is completed. (The part where bold is predicted)
| D1 | D2 | D3 | D4 | |
|---|---|---|---|---|
| U1 | 5.25 | 1.99 | 0. | 1.45 |
| U2 | 3.50 | 1.32 | 0. | 0.97 |
| U3 | 1.31 | 0.94 | 1.94 | 3.94 |
| U4 | 0.98 | 0.72 | 1.52 | 3.07 |
| U5 | 0. | 0.65 | 2.83 | 5.21 |
It is a difficult problem to decide which value k should be, and it is said that it depends on the input data (R).
- Increasing k decreases R-P × Q ^ T and increases the complexity of the model.
- Conversely, if k is made smaller, R-P × Q ^ T becomes larger and the complexity of the model becomes lower.
In this case, how can we determine which k is better? (If you know, please teach)
Link
-
scikit learn reference
- http://scikit-learn.org/stable/modules/generated/sklearn.decomposition.NMF.html
-
What is Matrix Factorization (easy-to-understand qiita)
- http://qiita.com/ysekky/items/c81ff24da0390a74fc6c
-
Matrix Factorization Techniques For Recommender Systems
- https://hpi.de/fileadmin/user_upload/fachgebiete/naumann/lehre/SS2011/Collaborative_Filtering/pres1-matrixfactorization.pdf
Recommended Posts