[PYTHON] Nowadays molecular competition (1)
Overview
Challenge molecular competitions for practicing RDKit and machine learning. Data assessment for the time being.
reference
Basically my work memo. Since it was last year's competition, there are already various summaries, so it might be better to look there.
- [Predicting Scalar Coupling Constants using Machine Learning] (https://medium.com/@liztersahakyan/predicting-scalar-coupling-constants-using-machine-learning-c213af14e862)
- [Review of Kaggle Molecular Competition-Overview of Competition / Application of GCN] (https://rishigami.hatenablog.com/entry/2019/08/29/090244)
Overview of the competition
https://www.kaggle.com/c/champs-scalar-coupling/ Find the value of magnetic interaction called scalar_coupling_constant
Number of data items and item name
For the time being, read it with pandas and nunique.
Test/training data
Although the number of molecules is 130,000, there is a value of the interaction required between each atom, so a total of 8 million records of test and training data.
- test.csv
--id: About 2.5 million
--molecule_name: about 45,000
- atom_index_0
- atom_index_1 --type: 8 types ('1JHC', '1JHN', '2JHC', '2JHH', '2JHN', '3JHC', '3JHH', '3JHN')
- train.csv
--id: About 4.7 million
--molecule_name: about 85,000
- atom_index_0
- atom_index_1 --type: 8 types --scalar_coupling_constant: Objective variable this time
Other data
molecule_name is the primary key
--dipole_moments.csv: Dipole moment (x, y, z data) --magnetic_shielding_tensors.csv: 8 combinations of X, Y, Z (XX, XY, ...) --potential energy: electron energy
molecule_name and atom_index are the primary keys
--mulliken_charges.csv: muliken charge (should be the one who simply halved the charge distribution between two points) - molecule_name --atom_index: 29 (Molecules with a maximum atomic number of 29) - mulliken_charge
--structures.csv (There are other xyz data) - molecule_name - atom_index --atom: 5 species (H, N, O, F, C) - x - y - z
Same as training data (4.7 million lines)
The sum of fc, sd, pso, and dso is the training data scalar_coupling_constant.
- scalar_coupling_contributions.csv
--molecule_name: 85,000
- atom_index_0
- atom_index_1
- type
- fc: Fermi Contact --sd: Spin-dipolar --pso: Paramagnetic spin-orbit --dso: Diamagnetic spin-orbit
Data distribution
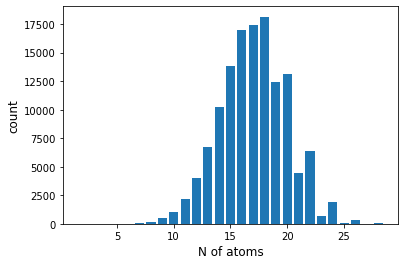
Number of molecules
You can see that there are many molecules of about 15 atoms (maybe it doesn't make much sense to see them)
path_structure = '../data/structures.csv'
df_structure = pd.read_csv(path_structure)
df_check = (
df_structure
.groupby('molecule_name')
.max('atom_index')
.reset_index()
.loc[:, ['molecule_name', 'atom_index']]
.groupby('atom_index').count()
)
plt.xlabel('N of atoms', fontsize=12)
plt.ylabel('count', fontsize=12)
plt.bar(df_check.index, df_check['molecule_name'])
plt.show()
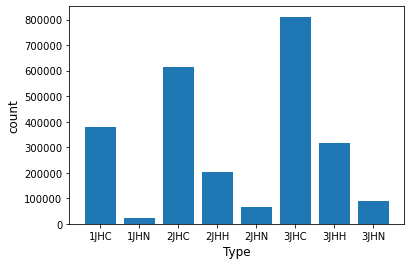
Test/training data
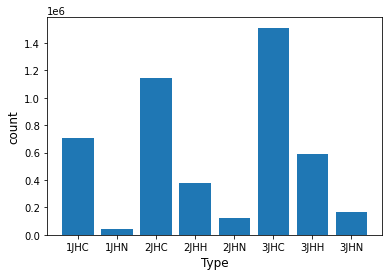
Check if the test data and training data have the same distribution by the number of 8 types of interactions → It seems to be divided randomly
[test data]
[Training data]
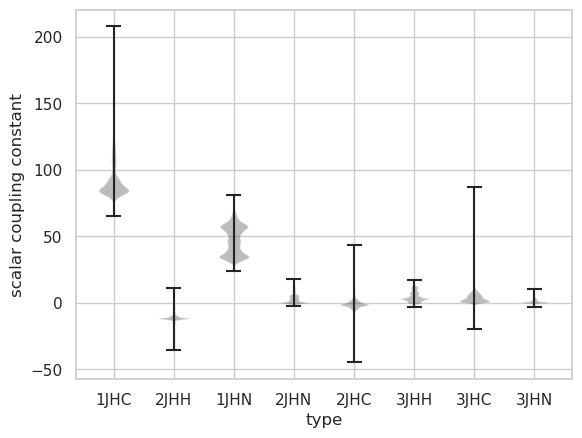
Distribution of values by type of interaction
1.1 Large variance of JHC 2. The value of 1JHC is about 80. Next, 1JHN is about 50. Others are around 0. 3.1 JHN has two peaks
Recommended Posts