[PYTHON] Correlation coefficient MIC and Hilbert-Schmidt independence test HSIC that can handle non-linear relationships
Pearson's correlation coefficient is a well-known index for measuring the independence of variables. However, it is an index that deals with linear (linear) relationships, and when there is a U-shaped relationship such as a quadratic function, it is judged as "no correlation". MIC and HSIC are known as indicators for dealing with such non-linear relationships, so I tried using the Python package.
All of the code below has been tested on Google Colaboratory.
A Python package that calculates the Maximal Information Coefficient (MIC)
You can pip install as follows:
!pip install minepy
After installation, prepare as follows.
from minepy import MINE
mine = MINE()
A Python package that calculates HSIC (Hilbert-Schmidt Independence Criterion)
Git clone as follows:
!git clone https://github.com/amber0309/HSIC.git
Prepare as follows.
from HSIC.HSIC import hsic_gam
Linear function
First, let's examine the linear function.
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
X = np.linspace(-1, 1, 51)
Y = X
plt.scatter(X, Y)

Pearson's correlation coefficient is
np.corrcoef(X, Y)[0, 1]
1.0
MIC is
mine.compute_score(X, Y)
mine.mic()
0.9999999999999998
HSSC is said to have lower independence as the next value is larger, and higher independence as the next value is smaller.
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
testStat / thresh
22.610487654066624
Let's add noise to the linear function.
import numpy as np
X = np.linspace(-1, 1, 51)
Y = X + np.random.rand(51,)
plt.scatter(X, Y)

Pearson's correlation coefficient is
np.corrcoef(X, Y)[0, 1]
0.9018483832673769
MIC is
mine.compute_score(X, Y)
mine.mic()
0.9997226475394071
HSIC
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
testStat / thresh
13.706487127525389
Let's calculate how Pearson's correlation coefficient changes as the amount of noise is increased little by little.
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = X + np.random.rand(51,) * d / 10
D.append(d)
C.append(np.corrcoef(X, Y)[0, 1])
plt.plot(D, C)

Similarly, let's calculate how the MIC changes as the noise level is increased little by little.
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = X + np.random.rand(51,) * d / 10
D.append(d)
mine.compute_score(X, Y)
C.append(mine.mic())
plt.plot(D, C)
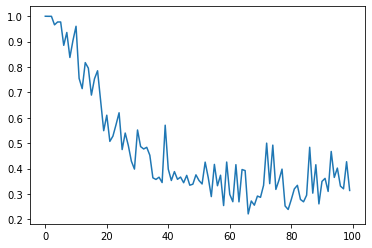
Similarly, let's calculate how the HSIC changes as the noise level is increased little by little.
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = X + np.random.rand(51,) * d / 10
D.append(d)
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
C.append(testStat / thresh)
plt.plot(D, C)

quadratic function
Next is the quadratic function.
import numpy as np
X = np.linspace(-1, 1, 51)
Y = X**2
plt.scatter(X, Y)

Pearson's correlation coefficient is
np.corrcoef(X, Y)[0, 1]
-2.3862043230836297e-17
MIC is
mine.compute_score(X, Y)
mine.mic()
0.9997226475394071
HSIC
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
testStat / thresh
9.285886928766184
Let's add noise.
X = np.linspace(-1, 1, 51)
Y = X**2 + np.random.rand(51,)
plt.scatter(X, Y)

Pearson's correlation coefficient is
np.corrcoef(X, Y)[0, 1]
0.04693076570744622
MIC is
mine.compute_score(X, Y)
mine.mic()
0.5071787519579662
HSIC
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
testStat / thresh
5.247036645612692
Let's calculate how Pearson's correlation coefficient changes as the amount of noise is increased little by little.
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = X**2 + np.random.rand(51,) * d / 10
D.append(d)
C.append(np.corrcoef(X, Y)[0, 1])
plt.plot(D, C)

Similarly, let's calculate how the MIC changes as the noise level is increased little by little.
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = X**2 + np.random.rand(51,) * d / 10
D.append(d)
mine.compute_score(X, Y)
C.append(mine.mic())
plt.plot(D, C)

Similarly, let's calculate how the HSIC changes as the noise level is increased little by little.
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = X**2 + np.random.rand(51,) * d / 10
D.append(d)
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
C.append(testStat / thresh)
plt.plot(D, C)

It can be seen that Pearson's correlation coefficient cannot detect the relationship of the quadratic function, while both MIC and HSIC can be detected. You can also see that the MIC will be largely unaffected by small noise.
Trigonometric function
Let's also look at trigonometric functions.
import numpy as np
X = np.linspace(-1, 1, 51)
Y = np.sin((X)*np.pi*2)
plt.scatter(X, Y)

Pearson's correlation coefficient is
np.corrcoef(X, Y)[0, 1]
-0.37651692742033543
MIC is
mine.compute_score(X, Y)
mine.mic()
0.9997226475394071
HSIC
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
testStat / thresh
3.212604441715373
Let's add noise.
import numpy as np
X = np.linspace(-1, 1, 51)
Y = np.sin((X+0.25)*np.pi*2) + np.random.rand(51,)
plt.scatter(X, Y)

Pearson's correlation coefficient is
np.corrcoef(X, Y)[0, 1]
0.049378342546505014
MIC is
mine.compute_score(X, Y)
mine.mic()
0.9997226475394071
HSIC
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
testStat / thresh
1.3135420547013614
Investigate the effect of increasing the noise. Pearson's correlation coefficient is
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = np.sin((X+0.25)*np.pi*2) + np.random.rand(51,) * d / 10
D.append(d)
C.append(np.corrcoef(X, Y)[0, 1])
plt.plot(D, C)

MIC is
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = np.sin((X+0.25)*np.pi*2) + np.random.rand(51,) * d / 10
D.append(d)
mine.compute_score(X, Y)
C.append(mine.mic())
plt.plot(D, C)

HSIC
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = np.sin((X+0.25)*np.pi*2) + np.random.rand(51,) * d / 10
D.append(d)
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
C.append(testStat / thresh)
plt.plot(D, C)

It was expected that Pearson's correlation coefficient could not detect the relationship of trigonometric functions with noise, but it seems that HSIC has also dropped out. Among them, I was surprised that MIC was able to detect the relationship.
Sigmoid function
Finally, there is the sigmoid function.
import numpy as np
X = np.linspace(-1, 1, 51)
Y = 1 / (1 + np.exp(-X*10))
plt.scatter(X, Y)

Pearson's correlation coefficient is
np.corrcoef(X, Y)[0, 1]
0.9354020629807919
MIC is
mine.compute_score(X, Y)
mine.mic()
0.9999999999999998
HSIC
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
testStat / thresh
27.433932271008874
Let's add noise.
import numpy as np
X = np.linspace(-1, 1, 51)
Y = 1 / (1 + np.exp(-X*10)) + np.random.rand(51,)
plt.scatter(X, Y)

Pearson's correlation coefficient is
np.corrcoef(X, Y)[0, 1]
0.7994507640548881
MIC is
mine.compute_score(X, Y)
mine.mic()
0.9997226475394071
HSIC
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
testStat / thresh
12.218008194240534
Let's see the effect of increasing the noise. Pearson's correlation coefficient is
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = 1 / (1 + np.exp(-X*10)) + np.random.rand(51,) * d / 10
D.append(d)
C.append(np.corrcoef(X, Y)[0, 1])
plt.plot(D, C)

MIC is
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = 1 / (1 + np.exp(-X*10)) + np.random.rand(51,) * d / 10
D.append(d)
mine.compute_score(X, Y)
C.append(mine.mic())
plt.plot(D, C)

HSIC
D = []
C = []
for d in range(100):
X = np.linspace(-1, 1, 51)
Y = 1 / (1 + np.exp(-X*10)) + np.random.rand(51,) * d / 10
D.append(d)
testStat, thresh = hsic_gam(X.reshape(len(X), 1), Y.reshape(len(Y), 1))
C.append(testStat / thresh)
plt.plot(D, C)

All of the indicators can detect the relationship, but in terms of noise resistance, MIC seems to be the strongest.
It is an individual impression
MIC MIC!